Grammatically Modeling and Predicting RNA Secondary Structures
Yasuo UEMURA (uemura-y@gold.cs.uec.ac.jp)
Aki HASEGAWA (haseg-ak@gold.cs.uec.ac.jp)
Satoshi KOBAYASHI (satoshi@cs.uec.ac.jp)
Takashi YOKOMORI (yokomori@cs.uec.ac.jp)
Department of Computer Science and Information Mathematics, University of Electro-Communications
1-5-1, Chofugaoka, Chofu, Tokyo 182, Japan
Abstract
Tree Adjunct Grammar for RNA (TAG
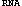 ) is a new grammatical device to model RNA secondary structures including pseudoknots. An efficient parsing algorithm for this grammar is developed, and applied to some computational problems concerning RNA secondary structures. With this parser, we first try to predict secondary structures of RNA sequences which are known to form pseudoknots structures, and show prediction results which nicely match the known structures. Further, a (-1) frameshift grammar is constructed based on a biological observation that a (-1) frameshift might be caused from some structural features of RNA sequences. The proposed grammar is used to find candidate sequences for (-1) frameshift in Human spumaretrovirus gag and pol genes.
) is a new grammatical device to model RNA secondary structures including pseudoknots. An efficient parsing algorithm for this grammar is developed, and applied to some computational problems concerning RNA secondary structures. With this parser, we first try to predict secondary structures of RNA sequences which are known to form pseudoknots structures, and show prediction results which nicely match the known structures. Further, a (-1) frameshift grammar is constructed based on a biological observation that a (-1) frameshift might be caused from some structural features of RNA sequences. The proposed grammar is used to find candidate sequences for (-1) frameshift in Human spumaretrovirus gag and pol genes.

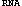 ) is a new grammatical device to model RNA secondary structures including pseudoknots. An efficient parsing algorithm for this grammar is developed, and applied to some computational problems concerning RNA secondary structures. With this parser, we first try to predict secondary structures of RNA sequences which are known to form pseudoknots structures, and show prediction results which nicely match the known structures. Further, a (-1) frameshift grammar is constructed based on a biological observation that a (-1) frameshift might be caused from some structural features of RNA sequences. The proposed grammar is used to find candidate sequences for (-1) frameshift in Human spumaretrovirus gag and pol genes.
) is a new grammatical device to model RNA secondary structures including pseudoknots. An efficient parsing algorithm for this grammar is developed, and applied to some computational problems concerning RNA secondary structures. With this parser, we first try to predict secondary structures of RNA sequences which are known to form pseudoknots structures, and show prediction results which nicely match the known structures. Further, a (-1) frameshift grammar is constructed based on a biological observation that a (-1) frameshift might be caused from some structural features of RNA sequences. The proposed grammar is used to find candidate sequences for (-1) frameshift in Human spumaretrovirus gag and pol genes.