Sequence Analysis of Zinc Finger
DNA-Binding Protein
Kotoko Nakata (nakata@nihs.nihs.go.jp)
Division of Chem-Bio Informatics
National Institute of Health Sciences
18-1, Kamiyoga 1-chome, Setagaya-ku, Tokyo 158
Abstract
Using the neural network algorithm with back-propagation traing procedure, we analysed the zinc finger DNA binding protein sequences. The patterns which were used in the neural network are amino acids sequence pattern, the electric charge and polarity, amino acids group properties, amino acids ancestral group, hydrophobicity, hydrophilicity and the secondary structure. For the comparison, the discriminant analysis was also tried. As for the TFIIIA type (Cys - X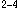 - Cys - X
- Cys - X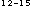 - His - X
- His - X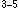 - His) (X is any amino acid) zinc finger DNA binding motifs, the prediction results reached high discrimination in the neural network algorithm and the discriminant analysis. Although each result of single perceptron algorithm is not always good in the case of the estrogen type (Cys - X
- His) (X is any amino acid) zinc finger DNA binding motifs, the prediction results reached high discrimination in the neural network algorithm and the discriminant analysis. Although each result of single perceptron algorithm is not always good in the case of the estrogen type (Cys - X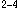 - Cys - X
- Cys - X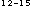 - Cys) zinc finge, the combination of the attributes reached high discrimination.
- Cys) zinc finge, the combination of the attributes reached high discrimination.
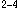 - Cys - X
- Cys - X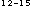 - His - X
- His - X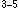 - His) (X is any amino acid) zinc finger DNA binding motifs, the prediction results reached high discrimination in the neural network algorithm and the discriminant analysis. Although each result of single perceptron algorithm is not always good in the case of the estrogen type (Cys - X
- His) (X is any amino acid) zinc finger DNA binding motifs, the prediction results reached high discrimination in the neural network algorithm and the discriminant analysis. Although each result of single perceptron algorithm is not always good in the case of the estrogen type (Cys - X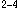 - Cys - X
- Cys - X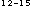 - Cys) zinc finge, the combination of the attributes reached high discrimination.
- Cys) zinc finge, the combination of the attributes reached high discrimination.